You can run this notebook in a live session or view it on Github.
Temporal and Spatial Smoothing#
This demo demonstrates climpred’s capabilities to postprocess decadal prediction output before skill verification.
Here, we showcase a set of methods to smooth out noise in the spatial and temporal domain.
# linting
%load_ext nb_black
%load_ext lab_black
%matplotlib inline
from climpred import PerfectModelEnsemble, HindcastEnsemble
from climpred.tutorial import load_dataset
import matplotlib.pylab as plt
import xarray as xr
# Sea surface temperature
v = "tos"
initialized3d = load_dataset("MPI-PM-DP-3D")[v]
control3d = load_dataset("MPI-control-3D")[v]
climpred requires that lead dimension has an attribute called units indicating what time units the lead is assocated with. Options are: years, seasons, months, weeks, pentads, days, hours, minutes, seconds. For the this data, the lead units are years.
initialized3d["lead"].attrs = {"units": "years"}
Temporal smoothing#
In order to reduce temporal noise, decadal predictions are recommended to take multi-year averages [Goddard et al., 2013].
pm = PerfectModelEnsemble(initialized3d).add_control(control3d)
/Users/aaron.spring/Coding/climpred/climpred/utils.py:191: UserWarning: Assuming annual resolution starting Jan 1st due to numeric inits. Please change ``init`` to a datetime if it is another resolution. We recommend using xr.CFTimeIndex as ``init``, see https://climpred.readthedocs.io/en/stable/setting-up-data.html.
warnings.warn(
/Users/aaron.spring/Coding/climpred/climpred/utils.py:191: UserWarning: Assuming annual resolution starting Jan 1st due to numeric inits. Please change ``init`` to a datetime if it is another resolution. We recommend using xr.CFTimeIndex as ``init``, see https://climpred.readthedocs.io/en/stable/setting-up-data.html.
warnings.warn(
PerfectModelEnsemble.smooth() ({"time":x}) aggregates over x timesteps in time dimensions lead and time. Here it does not matter whether you specify lead or time, temporal smoothing is applied to both time dimensions. Note that the time dimension labels are not changed by this temporal smoothing immediately.
pm_tsmoothed = pm.smooth({"lead": 3})
print("initialized", pm_tsmoothed.get_initialized().coords, "\n")
print("control", pm_tsmoothed.get_control().coords)
initialized Coordinates:
lon (y, x) float64 -47.25 -47.69 -48.12 -48.54 ... 131.3 132.5 133.8
lat (y, x) float64 76.36 76.3 76.24 76.17 ... -77.25 -77.39 -77.54
* lead (lead) int64 1 2 3
* init (init) object 3014-01-01 00:00:00 ... 3237-01-01 00:00:00
* member (member) int64 1 2 3 4
valid_time (lead, init) object 3015-01-01 00:00:00 ... 3240-01-01 00:00:00
control Coordinates:
lon (y, x) float64 -47.25 -47.69 -48.12 -48.54 ... 131.3 132.5 133.8
lat (y, x) float64 76.36 76.3 76.24 76.17 ... -77.25 -77.39 -77.54
* time (time) object 3000-01-01 00:00:00 ... 3047-01-01 00:00:00
But after calling PerfectModelEnsemble.verify(), the correct time aggregation label is automatically set.
pm_tsmoothed.verify(metric="rmse", comparison="m2e", dim=["init", "member"])[v].plot(
col="lead", vmin=0, vmax=0.7, yincrease=False, x="x"
)
<xarray.plot.facetgrid.FacetGrid at 0x14b302df0>
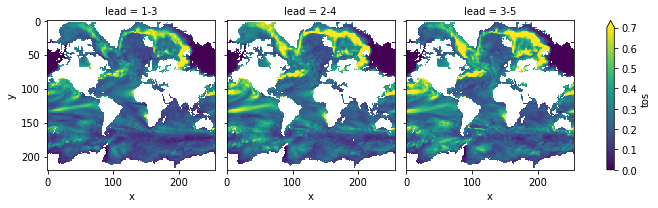
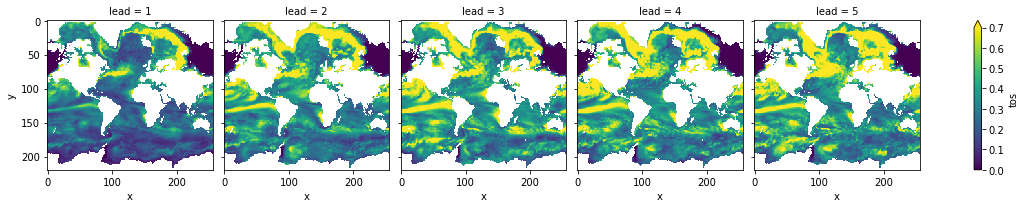
Compare to the prediction skill without smoothing (with rmse smaller is better):
pm.verify(metric="rmse", comparison="m2e", dim=["init", "member"])[v].plot(
col="lead", vmin=0, vmax=0.7, yincrease=False, x="x"
)
<xarray.plot.facetgrid.FacetGrid at 0x14b4f4970>
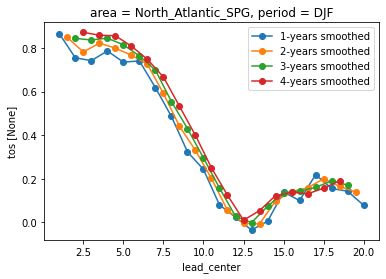
You can plot time-aggregated skill timeseries by specifying x=lead_center while plotting. Lead centers are placed in the center of the lead-aggregation boundaries. Note how the correlation of forecasts is increasing for longer time-aggregations because noise is smoothed out.
pm = PerfectModelEnsemble(load_dataset("MPI-PM-DP-1D"))
pm = pm.add_control(load_dataset("MPI-control-1D"))
pm_NA = pm.sel(area="North_Atlantic_SPG", period="DJF")[['tos']]
/Users/aaron.spring/Coding/climpred/climpred/utils.py:191: UserWarning: Assuming annual resolution starting Jan 1st due to numeric inits. Please change ``init`` to a datetime if it is another resolution. We recommend using xr.CFTimeIndex as ``init``, see https://climpred.readthedocs.io/en/stable/setting-up-data.html.
warnings.warn(
/Users/aaron.spring/Coding/climpred/climpred/utils.py:191: UserWarning: Assuming annual resolution starting Jan 1st due to numeric inits. Please change ``init`` to a datetime if it is another resolution. We recommend using xr.CFTimeIndex as ``init``, see https://climpred.readthedocs.io/en/stable/setting-up-data.html.
warnings.warn(
for time_agg in [1, 2, 3, 4]:
pm_NA.smooth({"lead": time_agg}).verify(
metric="acc", comparison="m2e", dim=["init", "member"]
).tos.plot(x="lead_center", marker="o", label=f"{time_agg}-years smoothed")
plt.legend()
<matplotlib.legend.Legend at 0x14b28d940>
Now, we showcase these smoothing features for HindcastEnsemble.smooth().
v = "SST"
initialized = load_dataset("CESM-DP-SST-3D")[v]
reconstruction = load_dataset("FOSI-SST-3D")[v]
# Move reconstruction into proper anomaly space
reconstruction = reconstruction - reconstruction.sel(time=slice(1964, 2014)).mean(
"time"
)
hindcast = HindcastEnsemble(initialized)
hindcast = hindcast.add_observations(reconstruction)
/Users/aaron.spring/Coding/climpred/climpred/utils.py:191: UserWarning: Assuming annual resolution starting Jan 1st due to numeric inits. Please change ``init`` to a datetime if it is another resolution. We recommend using xr.CFTimeIndex as ``init``, see https://climpred.readthedocs.io/en/stable/setting-up-data.html.
warnings.warn(
/Users/aaron.spring/Coding/climpred/climpred/utils.py:191: UserWarning: Assuming annual resolution starting Jan 1st due to numeric inits. Please change ``init`` to a datetime if it is another resolution. We recommend using xr.CFTimeIndex as ``init``, see https://climpred.readthedocs.io/en/stable/setting-up-data.html.
warnings.warn(
hindcast.smooth({"lead": 5}).verify(
metric="rmse", comparison="e2r", dim="init", alignment="same_verif"
)[v].plot(col="lead", robust=True)
<xarray.plot.facetgrid.FacetGrid at 0x14b1e1eb0>
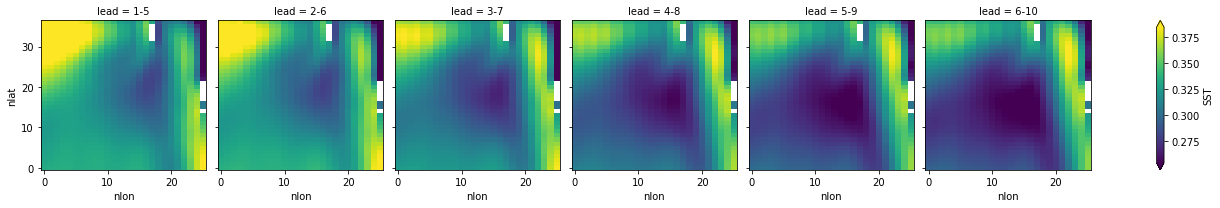
Spatial smoothing#
In order to reduce spatial noise, global decadal predictions are recommended to get regridded to a 5 degree longitude x 5 degree latitude grid as recommended [Goddard et al., 2013].
v = "tos"
pm_ssmoothed = pm.smooth({"lon": 5, "lat": 5})
pm_ssmoothed.get_initialized().coords
/Users/aaron.spring/anaconda3/envs/climpred-dev/lib/python3.9/site-packages/xarray/core/dataarray.py:784: FutureWarning: elementwise comparison failed; returning scalar instead, but in the future will perform elementwise comparison
return key in self.data
/Users/aaron.spring/anaconda3/envs/climpred-dev/lib/python3.9/site-packages/xesmf/backend.py:53: UserWarning: Latitude is outside of [-90, 90]
warnings.warn('Latitude is outside of [-90, 90]')
/Users/aaron.spring/anaconda3/envs/climpred-dev/lib/python3.9/site-packages/xarray/core/dataarray.py:784: FutureWarning: elementwise comparison failed; returning scalar instead, but in the future will perform elementwise comparison
return key in self.data
/Users/aaron.spring/anaconda3/envs/climpred-dev/lib/python3.9/site-packages/xesmf/backend.py:53: UserWarning: Latitude is outside of [-90, 90]
warnings.warn('Latitude is outside of [-90, 90]')
Coordinates:
* lead (lead) int64 1 2 3 4 5
* init (init) object 3014-01-01 00:00:00 ... 3237-01-01 00:00:00
* member (member) int64 1 2 3 4
valid_time (lead, init) object 3015-01-01 00:00:00 ... 3242-01-01 00:00:00
* lon (lon) float64 -180.0 -175.0 -170.0 -165.0 ... 170.0 175.0 180.0
* lat (lat) float64 -83.97 -78.97 -73.97 -68.97 ... 81.03 86.03 91.03
pm_ssmoothed.verify(metric="rmse", comparison="m2e", dim=["init", "member"])[v].T.plot(
col="lead", robust=True, yincrease=True,
)
<xarray.plot.facetgrid.FacetGrid at 0x149f67670>
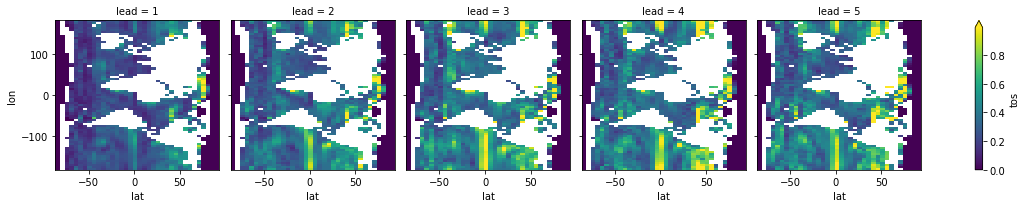
Spatial smoothing guesses the names corresponding to lon and lat in the coordinates of the PredictionEnsemble underlying datasets.
hindcast.get_initialized().coords
Coordinates:
TLAT (nlat, nlon) float64 -9.75 -9.75 -9.75 ... -0.1336 -0.1336
TLONG (nlat, nlon) float64 250.8 251.9 253.1 ... 276.7 277.8 278.9
* init (init) object 1954-01-01 00:00:00 ... 2017-01-01 00:00:00
* lead (lead) int32 1 2 3 4 5 6 7 8 9 10
TAREA (nlat, nlon) float64 3.661e+13 3.661e+13 ... 3.714e+13 3.714e+13
valid_time (lead, init) object 1955-01-01 00:00:00 ... 2027-01-01 00:00:00
hindcast.smooth({"lon": 1, "lat": 1}).get_initialized().coords
/Users/aaron.spring/anaconda3/envs/climpred-dev/lib/python3.9/site-packages/xarray/core/dataarray.py:784: FutureWarning: elementwise comparison failed; returning scalar instead, but in the future will perform elementwise comparison
return key in self.data
/Users/aaron.spring/anaconda3/envs/climpred-dev/lib/python3.9/site-packages/xarray/core/dataarray.py:784: FutureWarning: elementwise comparison failed; returning scalar instead, but in the future will perform elementwise comparison
return key in self.data
Coordinates:
* init (init) object 1954-01-01 00:00:00 ... 2017-01-01 00:00:00
* lead (lead) int32 1 2 3 4 5 6 7 8 9 10
valid_time (lead, init) object 1955-01-01 00:00:00 ... 2027-01-01 00:00:00
* lon (lon) float64 250.8 251.8 252.8 253.8 ... 277.8 278.8 279.8
* lat (lat) float64 -9.75 -8.75 -7.75 -6.75 ... -1.75 -0.7503 0.2497
HindcastEnsemble.smooth() ("goddard2013") creates 4-year means and 5x5 degree regridding as suggested in [Goddard et al., 2013].
pm.smooth("goddard2013").verify(
metric="acc", comparison="m2e", dim=["init", "member"]
).coords
/Users/aaron.spring/anaconda3/envs/climpred-dev/lib/python3.9/site-packages/xarray/core/dataarray.py:784: FutureWarning: elementwise comparison failed; returning scalar instead, but in the future will perform elementwise comparison
return key in self.data
/Users/aaron.spring/anaconda3/envs/climpred-dev/lib/python3.9/site-packages/xesmf/backend.py:53: UserWarning: Latitude is outside of [-90, 90]
warnings.warn('Latitude is outside of [-90, 90]')
/Users/aaron.spring/anaconda3/envs/climpred-dev/lib/python3.9/site-packages/xarray/core/dataarray.py:784: FutureWarning: elementwise comparison failed; returning scalar instead, but in the future will perform elementwise comparison
return key in self.data
/Users/aaron.spring/anaconda3/envs/climpred-dev/lib/python3.9/site-packages/xesmf/backend.py:53: UserWarning: Latitude is outside of [-90, 90]
warnings.warn('Latitude is outside of [-90, 90]')
Coordinates:
* lead (lead) <U3 '1-4' '2-5'
* lon (lon) float64 -180.0 -175.0 -170.0 -165.0 ... 170.0 175.0 180.0
* lat (lat) float64 -83.97 -78.97 -73.97 -68.97 ... 81.03 86.03 91.03
lead_center (lead) float64 2.5 3.5
References#
L. Goddard, A. Kumar, A. Solomon, D. Smith, G. Boer, P. Gonzalez, V. Kharin, W. Merryfield, C. Deser, S. J. Mason, B. P. Kirtman, R. Msadek, R. Sutton, E. Hawkins, T. Fricker, G. Hegerl, C. a. T. Ferro, D. B. Stephenson, G. A. Meehl, T. Stockdale, R. Burgman, A. M. Greene, Y. Kushnir, M. Newman, J. Carton, I. Fukumori, and T. Delworth. A verification framework for interannual-to-decadal predictions experiments. Climate Dynamics, 40(1-2):245–272, January 2013. doi:10/f4jjvf.