#!conda install intake fsspec intake-xarray -c conda-forge -y
#!pip install climetlab
#!pip install climetlab_s2s_ai_challenge
# linting
%load_ext nb_black
%load_ext lab_black
---------------------------------------------------------------------------
ModuleNotFoundError Traceback (most recent call last)
Cell In[3], line 2
1 # linting
----> 2 get_ipython().run_line_magic('load_ext', 'nb_black')
3 get_ipython().run_line_magic('load_ext', 'lab_black')
File ~/checkouts/readthedocs.org/user_builds/climpred/conda/latest/lib/python3.9/site-packages/IPython/core/interactiveshell.py:2456, in InteractiveShell.run_line_magic(self, magic_name, line, _stack_depth)
2454 kwargs['local_ns'] = self.get_local_scope(stack_depth)
2455 with self.builtin_trap:
-> 2456 result = fn(*args, **kwargs)
2458 # The code below prevents the output from being displayed
2459 # when using magics with decorator @output_can_be_silenced
2460 # when the last Python token in the expression is a ';'.
2461 if getattr(fn, magic.MAGIC_OUTPUT_CAN_BE_SILENCED, False):
File ~/checkouts/readthedocs.org/user_builds/climpred/conda/latest/lib/python3.9/site-packages/IPython/core/magics/extension.py:33, in ExtensionMagics.load_ext(self, module_str)
31 if not module_str:
32 raise UsageError('Missing module name.')
---> 33 res = self.shell.extension_manager.load_extension(module_str)
35 if res == 'already loaded':
36 print("The %s extension is already loaded. To reload it, use:" % module_str)
File ~/checkouts/readthedocs.org/user_builds/climpred/conda/latest/lib/python3.9/site-packages/IPython/core/extensions.py:76, in ExtensionManager.load_extension(self, module_str)
69 """Load an IPython extension by its module name.
70
71 Returns the string "already loaded" if the extension is already loaded,
72 "no load function" if the module doesn't have a load_ipython_extension
73 function, or None if it succeeded.
74 """
75 try:
---> 76 return self._load_extension(module_str)
77 except ModuleNotFoundError:
78 if module_str in BUILTINS_EXTS:
File ~/checkouts/readthedocs.org/user_builds/climpred/conda/latest/lib/python3.9/site-packages/IPython/core/extensions.py:91, in ExtensionManager._load_extension(self, module_str)
89 with self.shell.builtin_trap:
90 if module_str not in sys.modules:
---> 91 mod = import_module(module_str)
92 mod = sys.modules[module_str]
93 if self._call_load_ipython_extension(mod):
File ~/checkouts/readthedocs.org/user_builds/climpred/conda/latest/lib/python3.9/importlib/__init__.py:127, in import_module(name, package)
125 break
126 level += 1
--> 127 return _bootstrap._gcd_import(name[level:], package, level)
File <frozen importlib._bootstrap>:1030, in _gcd_import(name, package, level)
File <frozen importlib._bootstrap>:1007, in _find_and_load(name, import_)
File <frozen importlib._bootstrap>:984, in _find_and_load_unlocked(name, import_)
ModuleNotFoundError: No module named 'nb_black'
Calculate skill of S2S model ECMWF for daily global reforecasts#
import climpred
import xarray as xr
Get hindcast#
S2S output is hosted on the ECMWF S3 cloud, see https://github.com/ecmwf-lab/climetlab-s2s-ai-challenge
Two ways to access:
direct access via
intake_xarrayand integrated caching viafsspecclimetlabwith integrated caching
Resources:
https://intake.readthedocs.io/en/latest/
https://intake-xarray.readthedocs.io/en/latest/
https://intake-thredds.readthedocs.io/en/latest/
https://filesystem-spec.readthedocs.io/en/latest/
https://climetlab.readthedocs.io/en/latest/
Hindcasts/Reforecasts are stored in netcdf or grib format.
For each initialization init (CF convention: standard_name=forecast_reference_time) in the year 2020 (see dates), reforecasts are available started on the same dayofyear from 2000-2019.
Here we get the reforecast skill for 2m temperature of forecasts initialized 2nd Jan 2000-2019.
dates = xr.cftime_range(start="2020-01-02", freq="7D", end="2020-12-31")
var = "t2m"
date = dates[0].strftime("%Y%m%d")
date
'20200102'
intake_xarray#
import intake
import fsspec
from aiohttp import ClientSession, ClientTimeout
timeout = ClientTimeout(total=600)
fsspec.config.conf["https"] = dict(client_kwargs={"timeout": timeout})
import intake_xarray
cache_path = "my_caching_folder"
fsspec.config.conf["simplecache"] = {"cache_storage": cache_path, "same_names": True}
forecast = (
intake_xarray.NetCDFSource(
f"simplecache::https://storage.ecmwf.europeanweather.cloud/s2s-ai-challenge/data/test-input/0.3.0/netcdf/ecmwf-forecast-{var}-{date}.nc"
)
.to_dask()
.compute()
)
climetlab#
climetlab wraps cdsapi to download from the Copernicus Climate Data Store (CDS) and from plug-in sources:
https://climetlab.readthedocs.io/en/latest/
https://github.com/ecmwf-lab/climetlab-s2s-ai-challenge
https://cds.climate.copernicus.eu/cdsapp#!/home
https://github.com/ecmwf/cdsapi/
import climetlab
forecast_climetlab = (
climetlab.load_dataset(
"s2s-ai-challenge-test-input",
origin="ecmwf",
date=[20200102, 20200206], # get to initializations/forecast_times
parameter=var,
format="netcdf",
)
.to_xarray()
.compute()
)
By downloading data from this dataset, you agree to the terms and conditions defined at https://apps.ecmwf.int/datasets/data/s2s/licence/. If you do not agree with such terms, do not download the data.
xr.testing.assert_equal(forecast_climetlab.sel(forecast_time="2020-01"), forecast)
forecast_climetlab.coords
Coordinates:
* realization (realization) int64 0 1 2 3 4 5 6 7 ... 44 45 46 47 48 49 50
* forecast_time (forecast_time) datetime64[ns] 2020-01-02 2020-02-06
* lead_time (lead_time) timedelta64[ns] 1 days 2 days ... 45 days 46 days
* latitude (latitude) float64 90.0 88.5 87.0 85.5 ... -87.0 -88.5 -90.0
* longitude (longitude) float64 0.0 1.5 3.0 4.5 ... 355.5 357.0 358.5
valid_time (forecast_time, lead_time) datetime64[ns] 2020-01-03 ... 2...
Get observations#
Choose from:
CPC
ERA5
CPC as observations#
compare against CPC observations: http://iridl.ldeo.columbia.edu/SOURCES/.NOAA/.NCEP/.CPC/.temperature/.daily/
cache = True
if not cache:
chunk_dim = "T"
grid = 1.5
tmin = xr.open_dataset(
f"http://iridl.ldeo.columbia.edu/SOURCES/.NOAA/.NCEP/.CPC/.temperature/.daily/.tmin/T/(0000%201%20Jan%202020)/(0000%2001%20Apr%202020)/RANGEEDGES/X/0/{grid}/358.5/GRID/Y/90/{grid}/-90/GRID/dods",
chunks={chunk_dim: "auto"},
).rename({"tmin": "t"})
tmax = xr.open_dataset(
f"http://iridl.ldeo.columbia.edu/SOURCES/.NOAA/.NCEP/.CPC/.temperature/.daily/.tmax/T/(0000%201%20Jan%202020)/(0000%2001%20Apr%202020)/RANGEEDGES/X/0/{grid}/358.5/GRID/Y/90/{grid}/-90/GRID/dods",
chunks={chunk_dim: "auto"},
).rename({"tmax": "t"})
t = (tmin + tmax) / 2
t["T"] = xr.cftime_range(start="2020-01-01", freq="1D", periods=t.T.size)
t = t.rename({"X": "longitude", "Y": "latitude", "T": "time"})
t["t"].attrs = tmin["t"].attrs
t["t"].attrs["long_name"] = "Daily Temperature"
t = t.rename({"t": "t2m"}) + 273.15
t["t2m"].attrs["units"] = "K"
t.to_netcdf("my_caching_folder/obs_CPC.nc")
else:
obs = xr.open_dataset("my_caching_folder/obs_CPC.nc").compute()
ERA5 as observations#
compare against ERA5 reanalysis fetched via https://github.com/ecmwf/cdsapi/ from https://cds.climate.copernicus.eu/cdsapp#!/dataset/reanalysis-era5-single-levels?tab=overview
forecast_times = forecast_climetlab.set_coords("valid_time").valid_time.values.flatten()
# takes a while, continue with obs from above
obs_era = (
climetlab.load_source(
"cds",
"reanalysis-era5-single-levels",
product_type="reanalysis",
time=["00:00"],
grid=[1.5, 1.5],
param="2t",
date=forecast_times,
)
.to_xarray()
.squeeze(drop=True)
)
Forecast verification#
with HindcastEnsemble
# PredictionEnsemble converts `lead_time` to `lead` due to standard_name 'forecast_period' and
# pd.Timedelta to `int` and set lead.attrs['units'] accordingly
forecast_climetlab.coords
Coordinates:
* realization (realization) int64 0 1 2 3 4 5 6 7 ... 44 45 46 47 48 49 50
* forecast_time (forecast_time) datetime64[ns] 2020-01-02 2020-02-06
* lead_time (lead_time) timedelta64[ns] 1 days 2 days ... 45 days 46 days
* latitude (latitude) float64 90.0 88.5 87.0 85.5 ... -87.0 -88.5 -90.0
* longitude (longitude) float64 0.0 1.5 3.0 4.5 ... 355.5 357.0 358.5
valid_time (forecast_time, lead_time) datetime64[ns] 2020-01-03 ... 2...
fct = (
climpred.HindcastEnsemble(
forecast_climetlab.drop_vars("valid_time").isel(lead_time=range(4))
)
.add_observations(obs)
.compute()
)
metric_kwargs = dict(
metric="rmse", comparison="e2o", alignment="same_inits", dim="init"
)
/Users/aaron.spring/Coding/climpred/climpred/checks.py:234: UserWarning: Did not find dimension "init", but renamed dimension forecast_time with CF-complying standard_name "forecast_reference_time" to init.
warnings.warn(
/Users/aaron.spring/Coding/climpred/climpred/checks.py:234: UserWarning: Did not find dimension "member", but renamed dimension realization with CF-complying standard_name "realization" to member.
warnings.warn(
/Users/aaron.spring/Coding/climpred/climpred/checks.py:234: UserWarning: Did not find dimension "lead", but renamed dimension lead_time with CF-complying standard_name "forecast_period" to lead.
warnings.warn(
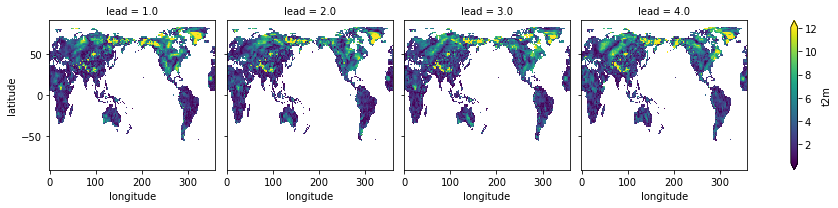
skill = fct.verify(**metric_kwargs)
skill[var].plot(col="lead", robust=True)
<xarray.plot.facetgrid.FacetGrid at 0x153344730>
# still experimental, help appreciated https://github.com/pangeo-data/climpred/issues/605
# with climpred.set_options(seasonality="month"):
# fct = fct.remove_bias(metric_kwargs["alignment"], cross_validate=False)
biweekly aggregates#
The s2s-ai-challenge requires biweekly target aggregates: https://s2s-ai-challenge.github.io/
There are two approaches possible for biweekly aggregates with climpred:
create biweekly aggregate timeseries for observations and forecasts and then use
climpreduse daily leads and daily observations and
HindcastEnsemble.smooth()to aggregate biweekly
biweekly aggregates before using climpred#
forecast#
from climpred.utils import convert_Timedelta_to_lead_units
forecast = convert_Timedelta_to_lead_units(
forecast_climetlab.rename({"lead_time": "lead"})
)
# create 14D averages
forecast_w12 = forecast.sel(lead=range(1, 14)).mean(dim="lead")
forecast_w34 = forecast.sel(lead=range(14, 28)).mean(dim="lead")
forecast_w56 = forecast.sel(lead=range(28, 42)).mean(dim="lead")
forecast_biweekly = xr.concat([forecast_w12, forecast_w34, forecast_w56], dim="lead")
forecast_biweekly["lead"] = [
1,
14,
28,
] # lead represents first day of biweekly aggregate
forecast_biweekly["lead"].attrs["units"] = "days"
forecast_biweekly.coords
Coordinates:
* realization (realization) int64 0 1 2 3 4 5 6 7 ... 44 45 46 47 48 49 50
* forecast_time (forecast_time) datetime64[ns] 2020-01-02 2020-02-06
* latitude (latitude) float64 90.0 88.5 87.0 85.5 ... -87.0 -88.5 -90.0
* longitude (longitude) float64 0.0 1.5 3.0 4.5 ... 355.5 357.0 358.5
* lead (lead) int64 1 14 28
observations#
Here I use rolling 14D mean, and set the time index to the starting date, i.e. I get 14D averages for every previous time date
# 14D rolling mean
obs_biweekly = obs.rolling(time=14, center=False).mean()
obs_biweekly = obs_biweekly.isel(time=slice(13, None)).assign_coords(
time=obs.time.isel(time=slice(None, -13))
) # time represents first day of the biweekly aggregate
fct_biweekly = climpred.HindcastEnsemble(forecast_biweekly).add_observations(
obs_biweekly
)
/Users/aaron.spring/Coding/climpred/climpred/checks.py:234: UserWarning: Did not find dimension "init", but renamed dimension forecast_time with CF-complying standard_name "forecast_reference_time" to init.
warnings.warn(
/Users/aaron.spring/Coding/climpred/climpred/checks.py:234: UserWarning: Did not find dimension "member", but renamed dimension realization with CF-complying standard_name "realization" to member.
warnings.warn(
obs_edges = (
obs_biweekly.groupby("time.month")
.quantile(q=[1 / 3, 2 / 3], dim="time", skipna=False)
.rename({"quantile": "category_edge"})
)
model_edges = (
forecast_biweekly.rename({"forecast_time": "init", "realization": "member"})
.groupby("init.month")
.quantile(q=[1 / 3, 2 / 3], dim=["init", "member"], skipna=False)
.rename({"quantile": "category_edge"})
)
rps#
verify with climpred.metrics._rps()
# use the same observational category_edges for observations and forecasts
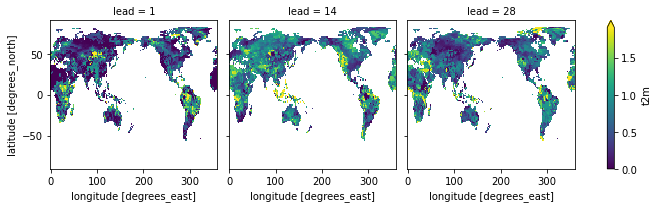
rps_same_edges = fct_biweekly.verify(
metric="rps",
comparison="m2o",
alignment="same_inits",
dim=["member", "init"],
category_edges=obs_edges,
)
rps_same_edges.t2m.plot(col="lead", robust=True)
<xarray.plot.facetgrid.FacetGrid at 0x15364c520>
# use different observational category_edges for observations and forecast category_edges for forecasts
rps_different_edges = fct_biweekly.verify(
metric="rps",
comparison="m2o",
alignment="same_inits",
dim=["member", "init"],
category_edges=(obs_edges, model_edges),
)
rps_different_edges.t2m.plot(col="lead", robust=True)
<xarray.plot.facetgrid.FacetGrid at 0x153619c70>
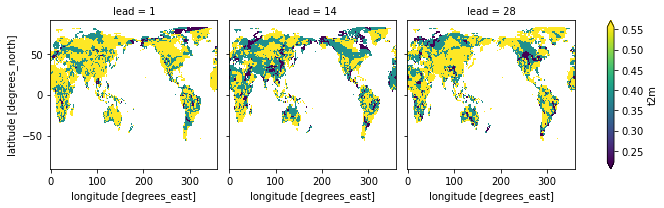
Using the model_edges to categorize the forecasts and obs_edges to categorize the observations separately is an inherent bias correction and improves rps.
biweekly aggregates with smooth#
climpred.classes.HindcastEnsemble.smooth()
fct = (
climpred.HindcastEnsemble(forecast_climetlab.drop_vars("valid_time")) # daily lead
.add_observations(obs)
.compute()
)
/Users/aaron.spring/Coding/climpred/climpred/checks.py:234: UserWarning: Did not find dimension "init", but renamed dimension forecast_time with CF-complying standard_name "forecast_reference_time" to init.
warnings.warn(
/Users/aaron.spring/Coding/climpred/climpred/checks.py:234: UserWarning: Did not find dimension "member", but renamed dimension realization with CF-complying standard_name "realization" to member.
warnings.warn(
/Users/aaron.spring/Coding/climpred/climpred/checks.py:234: UserWarning: Did not find dimension "lead", but renamed dimension lead_time with CF-complying standard_name "forecast_period" to lead.
warnings.warn(
# first smooth and then subselect leads
# use the same observational category_edges for observations and forecasts
fct.smooth(dict(lead=14), how="mean").verify(
metric="rps",
comparison="m2o",
alignment="same_inits",
dim=["member", "init"],
category_edges=obs_edges,
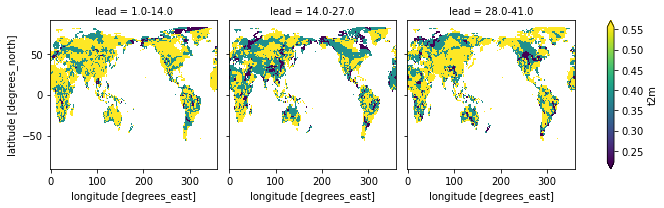
).sel(lead=["1.0-14.0", "14.0-27.0", "28.0-41.0"]).t2m.plot(col="lead", robust=True)
<xarray.plot.facetgrid.FacetGrid at 0x10f721550>
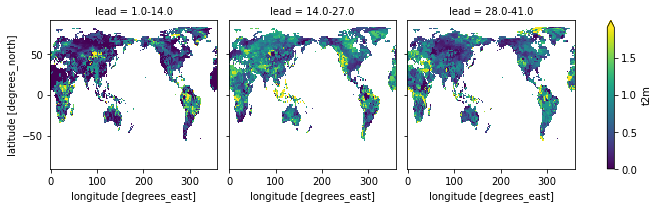
category_edges have to be smoothed before manually by climpred.smoothing.temporal_smoothing().
obs_edges = (
climpred.smoothing.temporal_smoothing(fct.get_observations(), dict(lead=14))
.groupby("time.month")
.quantile(q=[1 / 3, 2 / 3], dim="time", skipna=False)
.rename({"quantile": "category_edge"})
)
model_edges = (
climpred.smoothing.temporal_smoothing(fct.get_initialized(), dict(lead=14))
.groupby("init.month")
.quantile(q=[1 / 3, 2 / 3], dim=["init", "member"], skipna=False)
.rename({"quantile": "category_edge"})
)
# first smooth and then subselect leads
# use different observational category_edges for observations and forecast category_edges for forecasts
fct.smooth(dict(lead=14), how="mean").verify(
metric="rps",
comparison="m2o",
alignment="same_inits",
dim=["member", "init"],
category_edges=(obs_edges, model_edges),
).sel(lead=["1.0-14.0", "14.0-27.0", "28.0-41.0"]).t2m.plot(col="lead", robust=True)
<xarray.plot.facetgrid.FacetGrid at 0x14cc6f580>