Hindcast Predictions of Equatorial Pacific SSTs¶
In this example, we evaluate hindcasts (retrospective forecasts) of sea surface temperatures in the eastern equatorial Pacific from CESM-DPLE. These hindcasts are evaluated against a forced ocean–sea ice simulation that initializes the model.
See the quick start for an analysis of time series (rather than maps) from a hindcast prediction ensemble.
[1]:
import warnings
import cartopy.crs as ccrs # you may need to install cartopy, if not using climpred-dev
import cartopy.feature as cfeature
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
import climpred
from climpred import HindcastEnsemble
warnings.filterwarnings("ignore")
We’ll load in a small region of the eastern equatorial Pacific for this analysis example.
[2]:
hind = climpred.tutorial.load_dataset('CESM-DP-SST-3D')['SST']
recon = climpred.tutorial.load_dataset('FOSI-SST-3D')['SST']
hind
[2]:
<xarray.DataArray 'SST' (init: 64, lead: 10, nlat: 37, nlon: 26)>
[615680 values with dtype=float32]
Coordinates:
TLAT (nlat, nlon) float64 -9.75 -9.75 -9.75 ... -0.1336 -0.1336 -0.1336
TLONG (nlat, nlon) float64 250.8 251.9 253.1 254.2 ... 276.7 277.8 278.9
* init (init) float32 1954.0 1955.0 1956.0 1957.0 ... 2015.0 2016.0 2017.0
* lead (lead) int32 1 2 3 4 5 6 7 8 9 10
TAREA (nlat, nlon) float64 3.661e+13 3.661e+13 ... 3.714e+13 3.714e+13
Dimensions without coordinates: nlat, nlon- init: 64
- lead: 10
- nlat: 37
- nlon: 26
- ...
[615680 values with dtype=float32]
- TLAT(nlat, nlon)float64...
array([[-9.750341, -9.750341, -9.750341, ..., -9.750341, -9.750341, -9.750341], [-9.483209, -9.483209, -9.483209, ..., -9.483209, -9.483209, -9.483209], [-9.216077, -9.216077, -9.216077, ..., -9.216077, -9.216077, -9.216077], ..., [-0.667832, -0.667832, -0.667832, ..., -0.667832, -0.667832, -0.667832], [-0.400699, -0.400699, -0.400699, ..., -0.400699, -0.400699, -0.400699], [-0.133566, -0.133566, -0.133566, ..., -0.133566, -0.133566, -0.133566]]) - TLONG(nlat, nlon)float64...
array([[250.812507, 251.937507, 253.062507, ..., 276.687508, 277.812508, 278.937508], [250.812507, 251.937507, 253.062507, ..., 276.687508, 277.812508, 278.937508], [250.812507, 251.937507, 253.062507, ..., 276.687508, 277.812508, 278.937508], ..., [250.812507, 251.937507, 253.062507, ..., 276.687508, 277.812508, 278.937508], [250.812507, 251.937507, 253.062507, ..., 276.687508, 277.812508, 278.937508], [250.812507, 251.937507, 253.062507, ..., 276.687508, 277.812508, 278.937508]]) - init(init)float321954.0 1955.0 ... 2016.0 2017.0
- long_name :
- ensemble
- description :
- historical year corresponding to forecast year 1
- example :
- S=1955 for forecasts initialized on November 1 1954
array([1954., 1955., 1956., 1957., 1958., 1959., 1960., 1961., 1962., 1963., 1964., 1965., 1966., 1967., 1968., 1969., 1970., 1971., 1972., 1973., 1974., 1975., 1976., 1977., 1978., 1979., 1980., 1981., 1982., 1983., 1984., 1985., 1986., 1987., 1988., 1989., 1990., 1991., 1992., 1993., 1994., 1995., 1996., 1997., 1998., 1999., 2000., 2001., 2002., 2003., 2004., 2005., 2006., 2007., 2008., 2009., 2010., 2011., 2012., 2013., 2014., 2015., 2016., 2017.], dtype=float32) - lead(lead)int321 2 3 4 5 6 7 8 9 10
- long_name :
- forecast year
array([ 1, 2, 3, 4, 5, 6, 7, 8, 9, 10], dtype=int32)
- TAREA(nlat, nlon)float64...
- units :
- cm^2
- long_name :
- area of T cells
array([[3.660774e+13, 3.660774e+13, 3.660774e+13, ..., 3.660774e+13, 3.660774e+13, 3.660774e+13], [3.663666e+13, 3.663666e+13, 3.663666e+13, ..., 3.663666e+13, 3.663666e+13, 3.663666e+13], [3.666480e+13, 3.666480e+13, 3.666480e+13, ..., 3.666480e+13, 3.666480e+13, 3.666480e+13], ..., [3.714171e+13, 3.714171e+13, 3.714171e+13, ..., 3.714171e+13, 3.714171e+13, 3.714171e+13], [3.714332e+13, 3.714332e+13, 3.714332e+13, ..., 3.714332e+13, 3.714332e+13, 3.714332e+13], [3.714413e+13, 3.714413e+13, 3.714413e+13, ..., 3.714413e+13, 3.714413e+13, 3.714413e+13]])
These two example products cover a small portion of the eastern equatorial Pacific.
[3]:
ax = plt.axes(projection=ccrs.Orthographic(-80, 0))
p = ax.pcolormesh(recon.TLONG, recon.TLAT, recon.mean('time'),
transform=ccrs.PlateCarree(), cmap='twilight')
# ax.add_feature(cfeature.LAND, color='#d3d3d3')
ax.set_global()
plt.colorbar(p, label='Sea Surface Temperature [degC]')
ax.set(title='Example Data Coverage')
plt.show()
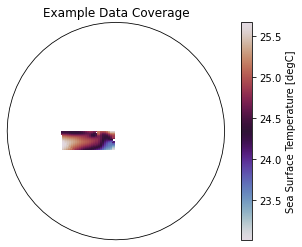
It is generally advisable to do all bias correction before instantiating a HindcastEnsemble. However, HindcastEnsemble can also be modified.
climpred requires that lead dimension has an attribute called units indicating what time units the lead is assocated with. Options are: years, seasons, months, weeks, pentads, days. For the this data, the lead units are years.
[4]:
hind['lead'].attrs['units'] = 'years'
[19]:
hindcast = HindcastEnsemble(hind)
hindcast = hindcast.add_observations(recon)
print(hindcast)
climpred.HindcastEnsemble
<Initialized Ensemble>
Dimensions: (init: 64, lead: 10, nlat: 37, nlon: 26)
Coordinates:
TLAT (nlat, nlon) float64 -9.75 -9.75 -9.75 ... -0.1336 -0.1336 -0.1336
TLONG (nlat, nlon) float64 250.8 251.9 253.1 254.2 ... 276.7 277.8 278.9
* init (init) object 1954-01-01 00:00:00 ... 2017-01-01 00:00:00
* lead (lead) int32 1 2 3 4 5 6 7 8 9 10
TAREA (nlat, nlon) float64 3.661e+13 3.661e+13 ... 3.714e+13 3.714e+13
Dimensions without coordinates: nlat, nlon
Data variables:
SST (init, lead, nlat, nlon) float32 ...- init: 64
- lead: 10
- nlat: 37
- nlon: 26
- TLAT(nlat, nlon)float64-9.75 -9.75 ... -0.1336 -0.1336
array([[-9.750341, -9.750341, -9.750341, ..., -9.750341, -9.750341, -9.750341], [-9.483209, -9.483209, -9.483209, ..., -9.483209, -9.483209, -9.483209], [-9.216077, -9.216077, -9.216077, ..., -9.216077, -9.216077, -9.216077], ..., [-0.667832, -0.667832, -0.667832, ..., -0.667832, -0.667832, -0.667832], [-0.400699, -0.400699, -0.400699, ..., -0.400699, -0.400699, -0.400699], [-0.133566, -0.133566, -0.133566, ..., -0.133566, -0.133566, -0.133566]]) - TLONG(nlat, nlon)float64250.8 251.9 253.1 ... 277.8 278.9
array([[250.812507, 251.937507, 253.062507, ..., 276.687508, 277.812508, 278.937508], [250.812507, 251.937507, 253.062507, ..., 276.687508, 277.812508, 278.937508], [250.812507, 251.937507, 253.062507, ..., 276.687508, 277.812508, 278.937508], ..., [250.812507, 251.937507, 253.062507, ..., 276.687508, 277.812508, 278.937508], [250.812507, 251.937507, 253.062507, ..., 276.687508, 277.812508, 278.937508], [250.812507, 251.937507, 253.062507, ..., 276.687508, 277.812508, 278.937508]]) - init(init)object1954-01-01 00:00:00 ... 2017-01-...
array([cftime.DatetimeProlepticGregorian(1954, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1955, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1956, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1957, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1958, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1959, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1960, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1961, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1962, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1963, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1964, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1965, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1966, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1967, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1968, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1969, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1970, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1971, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1972, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1973, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1974, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1975, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1976, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1977, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1978, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1979, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1980, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1981, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1982, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1983, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1984, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1985, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1986, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1987, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1988, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1989, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1990, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1991, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1992, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1993, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1994, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1995, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1996, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1997, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1998, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1999, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(2000, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(2001, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(2002, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(2003, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(2004, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(2005, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(2006, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(2007, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(2008, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(2009, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(2010, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(2011, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(2012, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(2013, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(2014, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(2015, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(2016, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(2017, 1, 1, 0, 0, 0, 0)], dtype=object) - lead(lead)int321 2 3 4 5 6 7 8 9 10
- long_name :
- forecast year
- units :
- years
array([ 1, 2, 3, 4, 5, 6, 7, 8, 9, 10], dtype=int32)
- TAREA(nlat, nlon)float643.661e+13 3.661e+13 ... 3.714e+13
- units :
- cm^2
- long_name :
- area of T cells
array([[3.660774e+13, 3.660774e+13, 3.660774e+13, ..., 3.660774e+13, 3.660774e+13, 3.660774e+13], [3.663666e+13, 3.663666e+13, 3.663666e+13, ..., 3.663666e+13, 3.663666e+13, 3.663666e+13], [3.666480e+13, 3.666480e+13, 3.666480e+13, ..., 3.666480e+13, 3.666480e+13, 3.666480e+13], ..., [3.714171e+13, 3.714171e+13, 3.714171e+13, ..., 3.714171e+13, 3.714171e+13, 3.714171e+13], [3.714332e+13, 3.714332e+13, 3.714332e+13, ..., 3.714332e+13, 3.714332e+13, 3.714332e+13], [3.714413e+13, 3.714413e+13, 3.714413e+13, ..., 3.714413e+13, 3.714413e+13, 3.714413e+13]])
- SST(init, lead, nlat, nlon)float32...
[615680 values with dtype=float32]
<Verification Data>
Dimensions: (nlat: 37, nlon: 26, time: 68)
Coordinates:
TLAT (nlat, nlon) float64 -9.75 -9.75 -9.75 ... -0.1336 -0.1336 -0.1336
TLONG (nlat, nlon) float64 250.8 251.9 253.1 254.2 ... 276.7 277.8 278.9
* time (time) object 1948-01-01 00:00:00 ... 2015-01-01 00:00:00
TAREA (nlat, nlon) float64 3.661e+13 3.661e+13 ... 3.714e+13 3.714e+13
Dimensions without coordinates: nlat, nlon
Data variables:
SST (time, nlat, nlon) float32 0.0029372794 0.0013880262 ... 1.4646177- nlat: 37
- nlon: 26
- time: 68
- TLAT(nlat, nlon)float64-9.75 -9.75 ... -0.1336 -0.1336
- units :
- degrees_north
- long_name :
- array of t-grid latitudes
array([[-9.75034113, -9.75034113, -9.75034113, -9.75034113, -9.75034113, -9.75034113, -9.75034113, -9.75034113, -9.75034113, -9.75034113, -9.75034113, -9.75034113, -9.75034113, -9.75034113, -9.75034113, -9.75034113, -9.75034113, -9.75034113, -9.75034113, -9.75034113, -9.75034113, -9.75034113, -9.75034113, -9.75034113, -9.75034113, -9.75034113], [-9.48320897, -9.48320897, -9.48320897, -9.48320897, -9.48320897, -9.48320897, -9.48320897, -9.48320897, -9.48320897, -9.48320897, -9.48320897, -9.48320897, -9.48320897, -9.48320897, -9.48320897, -9.48320897, -9.48320897, -9.48320897, -9.48320897, -9.48320897, -9.48320897, -9.48320897, -9.48320897, -9.48320897, -9.48320897, -9.48320897], [-9.21607677, -9.21607677, -9.21607677, -9.21607677, -9.21607677, -9.21607677, -9.21607677, -9.21607677, -9.21607677, -9.21607677, -9.21607677, -9.21607677, -9.21607677, -9.21607677, -9.21607677, -9.21607677, -9.21607677, -9.21607677, -9.21607677, -9.21607677, -9.21607677, -9.21607677, -9.21607677, -9.21607677, -9.21607677, -9.21607677], [-8.94894453, -8.94894453, -8.94894453, -8.94894453, -8.94894453, -8.94894453, -8.94894453, -8.94894453, -8.94894453, -8.94894453, ... -0.93496507, -0.93496507, -0.93496507, -0.93496507, -0.93496507, -0.93496507], [-0.6678322 , -0.6678322 , -0.6678322 , -0.6678322 , -0.6678322 , -0.6678322 , -0.6678322 , -0.6678322 , -0.6678322 , -0.6678322 , -0.6678322 , -0.6678322 , -0.6678322 , -0.6678322 , -0.6678322 , -0.6678322 , -0.6678322 , -0.6678322 , -0.6678322 , -0.6678322 , -0.6678322 , -0.6678322 , -0.6678322 , -0.6678322 , -0.6678322 , -0.6678322 ], [-0.40069932, -0.40069932, -0.40069932, -0.40069932, -0.40069932, -0.40069932, -0.40069932, -0.40069932, -0.40069932, -0.40069932, -0.40069932, -0.40069932, -0.40069932, -0.40069932, -0.40069932, -0.40069932, -0.40069932, -0.40069932, -0.40069932, -0.40069932, -0.40069932, -0.40069932, -0.40069932, -0.40069932, -0.40069932, -0.40069932], [-0.13356644, -0.13356644, -0.13356644, -0.13356644, -0.13356644, -0.13356644, -0.13356644, -0.13356644, -0.13356644, -0.13356644, -0.13356644, -0.13356644, -0.13356644, -0.13356644, -0.13356644, -0.13356644, -0.13356644, -0.13356644, -0.13356644, -0.13356644, -0.13356644, -0.13356644, -0.13356644, -0.13356644, -0.13356644, -0.13356644]]) - TLONG(nlat, nlon)float64250.8 251.9 253.1 ... 277.8 278.9
- units :
- degrees_east
- long_name :
- array of t-grid longitudes
array([[250.81250698, 251.93750701, 253.06250704, 254.18750707, 255.3125071 , 256.43750714, 257.56250717, 258.6875072 , 259.81250723, 260.93750726, 262.06250729, 263.18750732, 264.31250736, 265.43750739, 266.56250742, 267.68750745, 268.81250748, 269.93750751, 271.06250754, 272.18750757, 273.31250761, 274.43750764, 275.56250767, 276.6875077 , 277.81250773, 278.93750776], [250.81250698, 251.93750701, 253.06250704, 254.18750707, 255.3125071 , 256.43750714, 257.56250717, 258.6875072 , 259.81250723, 260.93750726, 262.06250729, 263.18750732, 264.31250736, 265.43750739, 266.56250742, 267.68750745, 268.81250748, 269.93750751, 271.06250754, 272.18750757, 273.31250761, 274.43750764, 275.56250767, 276.6875077 , 277.81250773, 278.93750776], [250.81250698, 251.93750701, 253.06250704, 254.18750707, 255.3125071 , 256.43750714, 257.56250717, 258.6875072 , 259.81250723, 260.93750726, 262.06250729, 263.18750732, 264.31250736, 265.43750739, 266.56250742, 267.68750745, 268.81250748, 269.93750751, 271.06250754, 272.18750757, 273.31250761, 274.43750764, 275.56250767, 276.6875077 , ... 255.3125071 , 256.43750714, 257.56250717, 258.6875072 , 259.81250723, 260.93750726, 262.06250729, 263.18750732, 264.31250736, 265.43750739, 266.56250742, 267.68750745, 268.81250748, 269.93750751, 271.06250754, 272.18750757, 273.31250761, 274.43750764, 275.56250767, 276.6875077 , 277.81250773, 278.93750776], [250.81250698, 251.93750701, 253.06250704, 254.18750707, 255.3125071 , 256.43750714, 257.56250717, 258.6875072 , 259.81250723, 260.93750726, 262.06250729, 263.18750732, 264.31250736, 265.43750739, 266.56250742, 267.68750745, 268.81250748, 269.93750751, 271.06250754, 272.18750757, 273.31250761, 274.43750764, 275.56250767, 276.6875077 , 277.81250773, 278.93750776], [250.81250698, 251.93750701, 253.06250704, 254.18750707, 255.3125071 , 256.43750714, 257.56250717, 258.6875072 , 259.81250723, 260.93750726, 262.06250729, 263.18750732, 264.31250736, 265.43750739, 266.56250742, 267.68750745, 268.81250748, 269.93750751, 271.06250754, 272.18750757, 273.31250761, 274.43750764, 275.56250767, 276.6875077 , 277.81250773, 278.93750776]]) - time(time)object1948-01-01 00:00:00 ... 2015-01-...
array([cftime.DatetimeProlepticGregorian(1948, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1949, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1950, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1951, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1952, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1953, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1954, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1955, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1956, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1957, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1958, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1959, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1960, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1961, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1962, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1963, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1964, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1965, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1966, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1967, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1968, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1969, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1970, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1971, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1972, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1973, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1974, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1975, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1976, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1977, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1978, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1979, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1980, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1981, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1982, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1983, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1984, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1985, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1986, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1987, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1988, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1989, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1990, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1991, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1992, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1993, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1994, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1995, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1996, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1997, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1998, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1999, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(2000, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(2001, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(2002, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(2003, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(2004, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(2005, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(2006, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(2007, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(2008, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(2009, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(2010, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(2011, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(2012, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(2013, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(2014, 1, 1, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(2015, 1, 1, 0, 0, 0, 0)], dtype=object) - TAREA(nlat, nlon)float643.661e+13 3.661e+13 ... 3.714e+13
- units :
- cm^2
- long_name :
- area of T cells
array([[3.66077352e+13, 3.66077352e+13, 3.66077352e+13, 3.66077352e+13, 3.66077352e+13, 3.66077352e+13, 3.66077352e+13, 3.66077352e+13, 3.66077352e+13, 3.66077352e+13, 3.66077352e+13, 3.66077352e+13, 3.66077352e+13, 3.66077352e+13, 3.66077352e+13, 3.66077352e+13, 3.66077352e+13, 3.66077352e+13, 3.66077352e+13, 3.66077352e+13, 3.66077352e+13, 3.66077352e+13, 3.66077352e+13, 3.66077352e+13, 3.66077352e+13, 3.66077352e+13], [3.66366633e+13, 3.66366633e+13, 3.66366633e+13, 3.66366633e+13, 3.66366633e+13, 3.66366633e+13, 3.66366633e+13, 3.66366633e+13, 3.66366633e+13, 3.66366633e+13, 3.66366633e+13, 3.66366633e+13, 3.66366633e+13, 3.66366633e+13, 3.66366633e+13, 3.66366633e+13, 3.66366633e+13, 3.66366633e+13, 3.66366633e+13, 3.66366633e+13, 3.66366633e+13, 3.66366633e+13, 3.66366633e+13, 3.66366633e+13, 3.66366633e+13, 3.66366633e+13], [3.66647951e+13, 3.66647951e+13, 3.66647951e+13, 3.66647951e+13, 3.66647951e+13, 3.66647951e+13, 3.66647951e+13, 3.66647951e+13, 3.66647951e+13, 3.66647951e+13, 3.66647951e+13, 3.66647951e+13, 3.66647951e+13, 3.66647951e+13, 3.66647951e+13, 3.66647951e+13, 3.66647951e+13, 3.66647951e+13, 3.66647951e+13, 3.66647951e+13, 3.66647951e+13, 3.66647951e+13, 3.66647951e+13, 3.66647951e+13, ... 3.71417082e+13, 3.71417082e+13, 3.71417082e+13, 3.71417082e+13, 3.71417082e+13, 3.71417082e+13, 3.71417082e+13, 3.71417082e+13, 3.71417082e+13, 3.71417082e+13, 3.71417082e+13, 3.71417082e+13, 3.71417082e+13, 3.71417082e+13, 3.71417082e+13, 3.71417082e+13, 3.71417082e+13, 3.71417082e+13, 3.71417082e+13, 3.71417082e+13, 3.71417082e+13, 3.71417082e+13], [3.71433228e+13, 3.71433228e+13, 3.71433228e+13, 3.71433228e+13, 3.71433228e+13, 3.71433228e+13, 3.71433228e+13, 3.71433228e+13, 3.71433228e+13, 3.71433228e+13, 3.71433228e+13, 3.71433228e+13, 3.71433228e+13, 3.71433228e+13, 3.71433228e+13, 3.71433228e+13, 3.71433228e+13, 3.71433228e+13, 3.71433228e+13, 3.71433228e+13, 3.71433228e+13, 3.71433228e+13, 3.71433228e+13, 3.71433228e+13, 3.71433228e+13, 3.71433228e+13], [3.71441302e+13, 3.71441302e+13, 3.71441302e+13, 3.71441302e+13, 3.71441302e+13, 3.71441302e+13, 3.71441302e+13, 3.71441302e+13, 3.71441302e+13, 3.71441302e+13, 3.71441302e+13, 3.71441302e+13, 3.71441302e+13, 3.71441302e+13, 3.71441302e+13, 3.71441302e+13, 3.71441302e+13, 3.71441302e+13, 3.71441302e+13, 3.71441302e+13, 3.71441302e+13, 3.71441302e+13, 3.71441302e+13, 3.71441302e+13, 3.71441302e+13, 3.71441302e+13]])
- SST(time, nlat, nlon)float320.0029372794 ... 1.4646177
array([[[ 2.93727941e-03, 1.38802617e-03, 2.92755570e-03, ..., 1.95969343e-01, 2.05750450e-01, 2.24518195e-01], [ 2.77975504e-03, 2.61594728e-03, 4.65755817e-03, ..., 2.09077492e-01, 2.11955756e-01, 2.27542236e-01], [ 3.50267743e-03, 4.17638291e-03, 6.56097988e-03, ..., 2.20962301e-01, 2.16155961e-01, 2.29898304e-01], ..., [ 5.97333550e-01, 5.78063488e-01, 5.88940442e-01, ..., 3.31806272e-01, 2.79064924e-01, 2.32759699e-01], [ 6.26646996e-01, 6.15216076e-01, 6.28627181e-01, ..., 3.01406294e-01, 2.52097368e-01, 2.16521353e-01], [ 6.38442695e-01, 6.34791136e-01, 6.49721205e-01, ..., 2.65346617e-01, 2.17654392e-01, 1.95610940e-01]], [[-2.49124572e-01, -2.59037554e-01, -2.65844584e-01, ..., 7.19745085e-02, 1.38766274e-01, 1.87652960e-01], [-2.58721560e-01, -2.67041177e-01, -2.72869289e-01, ..., 8.51150975e-02, 1.53196067e-01, 2.06883743e-01], [-2.67771900e-01, -2.74739027e-01, -2.79625237e-01, ..., 9.69655663e-02, 1.65454820e-01, 2.26623386e-01], ... 4.64634031e-01, 5.57793438e-01, 6.71379328e-01], [ 7.51996040e-01, 7.67355740e-01, 7.84627318e-01, ..., 4.75749403e-01, 5.55110216e-01, 6.52999043e-01], [ 7.87040412e-01, 8.05157423e-01, 8.20713103e-01, ..., 4.87379164e-01, 5.49887836e-01, 6.27978265e-01]], [[ 1.85323671e-01, 1.99588254e-01, 2.18181387e-01, ..., 1.39085460e+00, 1.47076893e+00, 1.54842031e+00], [ 2.27164060e-01, 2.42421165e-01, 2.62142003e-01, ..., 1.43521655e+00, 1.51577163e+00, 1.57986200e+00], [ 2.70851910e-01, 2.86998034e-01, 3.07614982e-01, ..., 1.47523284e+00, 1.55554390e+00, 1.60825145e+00], ..., [ 2.24369025e+00, 2.22937536e+00, 2.23616290e+00, ..., 1.72568142e+00, 1.60193706e+00, 1.55987763e+00], [ 2.29228115e+00, 2.27476192e+00, 2.27866101e+00, ..., 1.70129907e+00, 1.56179929e+00, 1.51788628e+00], [ 2.32319808e+00, 2.30612445e+00, 2.30154037e+00, ..., 1.67635739e+00, 1.51848137e+00, 1.46461773e+00]]], dtype=float32)
[6]:
# Quick look at the HindEnsemble timeseries: only works for 1-dimensional data, therefore take spatial mean
hindcast.mean(['nlat', 'nlon']).plot()
[6]:
<AxesSubplot:xlabel='time', ylabel='SST'>
We first need to remove the same climatology that was used to drift-correct the CESM-DPLE.
[10]:
recon = recon - recon.sel(time=slice(1964,2014)).mean('time')
hindcast = hindcast.add_observations(recon)
hindcast.mean(['nlat', 'nlon']).plot()
[10]:
<AxesSubplot:xlabel='time', ylabel='SST'>
We’ll also detrend the reconstruction over its time dimension and initialized forecast ensemble over lead.
[20]:
from esmtools.stats import rm_trend
hindcast_detrend = hindcast.map(rm_trend, dim='lead').map(rm_trend, dim='time')
[21]:
hindcast_detrend.mean(['nlat', 'nlon']).plot()
[21]:
<AxesSubplot:xlabel='time', ylabel='SST'>
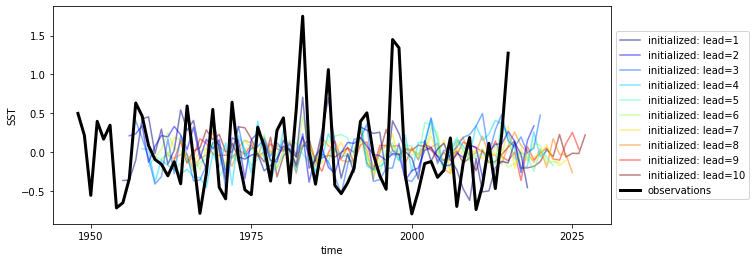
Although functions can be called directly in climpred, we suggest that you use our classes (HindcastEnsemble and PerfectModelEnsemble) to make analysis code cleaner.
Anomaly Correlation Coefficient of SSTs¶
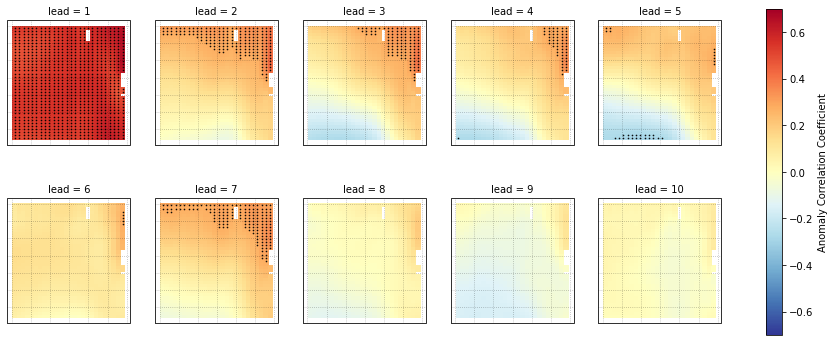
We can now compute the ACC over all leads and all grid cells.
[13]:
predictability = hindcast_detrend.verify(metric='acc', comparison='e2o', dim='init', alignment='same_verif')
print(predictability)
<xarray.Dataset>
Dimensions: (lead: 10, nlat: 37, nlon: 26, skill: 1)
Coordinates:
* lead (lead) int32 1 2 3 4 5 6 7 8 9 10
TLONG (nlat, nlon) float64 250.8 251.9 253.1 254.2 ... 276.7 277.8 278.9
TAREA (nlat, nlon) float64 3.661e+13 3.661e+13 ... 3.714e+13 3.714e+13
* skill (skill) <U4 'init'
Dimensions without coordinates: nlat, nlon
Data variables:
SST (lead, nlat, nlon) float64 0.5079 0.5049 0.5024 ... 0.0701 0.08838
We use the pval keyword to get associated p-values for our ACCs. We can then mask our final maps based on 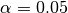 .
.
[14]:
significance = hindcast_detrend.verify(metric='p_pval', comparison='e2o', dim='init', alignment='same_verif')
# FIX: add missing coords to results
significance.coords['TLAT'] = hind['TLAT']
predictability.coords['TLAT'] = hind['TLAT']
[15]:
# Mask latitude and longitude by significance for stippling.
siglat = significance.TLAT.where(significance.SST <= 0.05)
siglon = significance.TLONG.where(significance.SST <= 0.05)
[16]:
p = predictability.SST.plot.pcolormesh(x='TLONG', y='TLAT',
transform=ccrs.PlateCarree(),
col='lead', col_wrap=5,
subplot_kws={'projection': ccrs.PlateCarree(),
'aspect': 3},
cbar_kwargs={'label': 'Anomaly Correlation Coefficient'},
vmin=-0.7, vmax=0.7,
cmap='RdYlBu_r')
for i, ax in enumerate(p.axes.flat):
# ax.add_feature(cfeature.LAND, color='#d3d3d3', zorder=4)
ax.gridlines(alpha=0.3, color='k', linestyle=':')
# Add significance stippling
ax.scatter(siglon.isel(lead=i),
siglat.isel(lead=i),
color='k',
marker='.',
s=1.5,
transform=ccrs.PlateCarree())
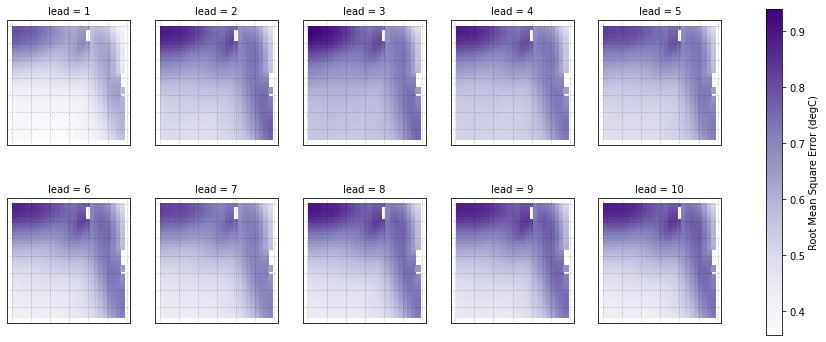
Root Mean Square Error of SSTs¶
We can also check error in our forecasts, just by changing the metric keyword.
[17]:
rmse = hindcast_detrend.verify(metric='rmse', comparison='e2o', dim='init', alignment='same_verif')
rmse.coords['TLAT'] = hind['TLAT']
[18]:
p = rmse.SST.plot.pcolormesh(x='TLONG', y='TLAT',
transform=ccrs.PlateCarree(),
col='lead', col_wrap=5,
subplot_kws={'projection': ccrs.PlateCarree(),
'aspect': 3},
cbar_kwargs={'label': 'Root Mean Square Error (degC)'},
cmap='Purples')
for ax in p.axes.flat:
# ax.add_feature(cfeature.LAND, color='#d3d3d3', zorder=4)
ax.gridlines(alpha=0.3, color='k', linestyle=':')